[1] "/Library/Frameworks/R.framework/Versions/4.5-arm64/Resources/library"R Packages
Duke University
Package logistics
What are packages?
R packages are just a collection of files - R code, compiled code (C, C++, etc.), data, documentation, and others that live in your library path.
[1] "abind" "airports" "AnnotationDbi"
[4] "ape" "AsioHeaders" "askpass"
[7] "backports" "base" "base64enc"
[10] "bench" "bestridge" "BH"
[13] "bigD" "Biobase" "BiocGenerics"
[16] "BiocManager" "BiocVersion" "Biostrings"
[19] "bit" "bit64" "bitops"
[22] "blob" "boot" "brew"
[25] "brio" "broom" "bslib"
[28] "cachem" "callr" "car"
[31] "carData" "cards" "cardx"
[34] "caret" "castor" "cellranger"
[37] "checkmate" "cherryblossom" "chromote"
[40] "chron" "class" "classInt"
[43] "cli" "clipr" "clock"
[46] "cluster" "clusterGeneration" "coda"
[49] "codetools" "colorspace" "combinat"
[52] "commonmark" "compiler" "conflicted"
[55] "corrplot" "countdown" "cowplot"
[58] "cpp11" "crayon" "credentials"
[61] "crosstalk" "curl" "data.table"
[64] "datasets" "datawizard" "DBI"
[67] "dbplyr" "DelayedArray" "dendextend"
[70] "DEoptim" "Deriv" "desc"
[73] "deSolve" "devEMF" "devtools"
[76] "diagram" "DiagrammeR" "dials"
[79] "DiceDesign" "diffobj" "digest"
[82] "distr" "distributional" "doBy"
[85] "doParallel" "dotCall64" "downlit"
[88] "dplyr" "DSA.CountData" "DT"
[91] "dtplyr" "duckdb" "e1071"
[94] "editData" "ellipse" "ellipsis"
[97] "emmeans" "estimability" "evaluate"
[100] "expm" "eyedata" "factoextra"
[103] "FactoMineR" "fansi" "faraway"
[106] "farver" "fastmap" "fastmatch"
[109] "fftwtools" "fields" "flare"
[112] "flashClust" "flextable" "fontawesome"
[115] "fontBitstreamVera" "fontLiberation" "fontquiver"
[118] "forcats" "foreach" "foreign"
[121] "Formula" "fs" "furrr"
[124] "future" "future.apply" "gapminder"
[127] "gargle" "gdtools" "geiger"
[130] "gender" "generics" "GenomeInfoDb"
[133] "GenomeInfoDbData" "GenomicRanges" "GEOquery"
[136] "gert" "GGally" "gganimate"
[139] "ggforce" "ggplot2" "ggpubr"
[142] "ggrepel" "ggridges" "ggsci"
[145] "ggsignif" "ggstats" "gh"
[148] "ghclass" "gifski" "gitcreds"
[151] "glmnet" "globals" "glue"
[154] "googledrive" "googlesheets4" "gower"
[157] "GPArotation" "GPBayes" "GPfit"
[160] "graphics" "grDevices" "grid"
[163] "gridExtra" "gsubfn" "gt"
[166] "gtable" "gtools" "gtsummary"
[169] "hardhat" "haven" "here"
[172] "hgu133a.db" "hgu95av2.db" "highr"
[175] "Hmisc" "hms" "htmlTable"
[178] "htmltools" "htmlwidgets" "httpuv"
[181] "httr" "httr2" "ids"
[184] "igraph" "infer" "ini"
[187] "inline" "insight" "ipred"
[190] "IRanges" "isoband" "iterators"
[193] "janeaustenr" "janitor" "jquerylib"
[196] "jsonlite" "juicyjuice" "kableExtra"
[199] "keep" "KEGGREST" "KernSmooth"
[202] "knitr" "labeling" "later"
[205] "latex2exp" "lattice" "lava"
[208] "lazyeval" "leaps" "lhs"
[211] "lifecycle" "limma" "listenv"
[214] "litedown" "lme4" "lmtest"
[217] "lobstr" "loo" "lpSolve"
[220] "lubridate" "magick" "magrittr"
[223] "maps" "markdown" "MASS"
[226] "Matrix" "MatrixGenerics" "MatrixModels"
[229] "matrixsampling" "matrixStats" "mcmcse"
[232] "memoise" "methods" "mgcv"
[235] "microbenchmark" "mime" "miniUI"
[238] "minqa" "mitools" "mlbench"
[241] "mnormt" "modeldata" "modelenv"
[244] "ModelMetrics" "modelr" "moonBook"
[247] "multcomp" "multcompView" "mvtnorm"
[250] "naturalsort" "ncbit" "nlme"
[253] "nloptr" "NLP" "nnet"
[256] "nortest" "numDeriv" "nycflights13"
[259] "officer" "openintro" "openNLP"
[262] "openNLPdata" "openssl" "openxlsx"
[265] "optimParallel" "org.Hs.eg.db" "org.Rn.eg.db"
[268] "otel" "ouch" "palmerpenguins"
[271] "parallel" "parallelly" "parsnip"
[274] "patchwork" "pbkrtest" "phangorn"
[277] "pheatmap" "phytools" "pillar"
[280] "pkgbuild" "pkgconfig" "pkgdown"
[283] "pkgload" "plogr" "plotly"
[286] "plotrix" "plsgenomics" "plyr"
[289] "png" "polspline" "polyclip"
[292] "polynom" "pomp" "posterior"
[295] "praise" "prettyunits" "prismatic"
[298] "pROC" "processx" "prodlim"
[301] "profmem" "profvis" "progress"
[304] "progressr" "promises" "proto"
[307] "proxy" "pryr" "ps"
[310] "psych" "purrr" "qdap"
[313] "qdapDictionaries" "qdapRegex" "qdapTools"
[316] "quadprog" "quantreg" "queryparser"
[319] "QuickJSR" "R.methodsS3" "R.oo"
[322] "R.utils" "R6" "ragg"
[325] "randomForest" "rappdirs" "rat2302.db"
[328] "rbibutils" "rcmdcheck" "RColorBrewer"
[331] "Rcpp" "RcppArmadillo" "RcppEigen"
[334] "RcppParallel" "RcppProgress" "RcppTOML"
[337] "RCurl" "Rdpack" "reactable"
[340] "reactablefmtr" "reactR" "readr"
[343] "readxl" "recipes" "reformulas"
[346] "rematch" "rematch2" "remotes"
[349] "rentrez" "reprex" "repurrrsive"
[352] "reshape2" "reticulate" "rgl"
[355] "RhpcBLASctl" "ridge" "rio"
[358] "rJava" "rlang" "rmarkdown"
[361] "rms" "roxygen2" "rpart"
[364] "rprojroot" "RRphylo" "rrtable"
[367] "rsample" "RSpectra" "RSQLite"
[370] "rstan" "rstatix" "rstudioapi"
[373] "rversions" "rvest" "rvg"
[376] "s2" "S4Arrays" "S4Vectors"
[379] "S7" "sandwich" "sass"
[382] "scales" "scatterplot3d" "selectr"
[385] "sessioninfo" "sf" "sfd"
[388] "sfsmisc" "shape" "shiny"
[391] "shinycssloaders" "shinyWidgets" "sjlabelled"
[394] "sjmisc" "slam" "slider"
[397] "sloop" "snakecase" "SnowballC"
[400] "sourcetools" "spam" "SparseArray"
[403] "SparseM" "sparsevctrs" "spatial"
[406] "splines" "sqldf" "SQUAREM"
[409] "srvyr" "srvyrexploR" "StanHeaders"
[412] "startupmsg" "statmod" "stats"
[415] "stats4" "stringdist" "stringi"
[418] "stringr" "subplex" "SummarizedExperiment"
[421] "survey" "survival" "svglite"
[424] "sys" "systemfonts" "tailor"
[427] "tcltk" "TeachingDemos" "tensorA"
[430] "testthat" "textshaping" "TH.data"
[433] "tibble" "tidymodels" "tidyquery"
[436] "tidyr" "tidyselect" "tidytext"
[439] "tidyverse" "timechange" "timeDate"
[442] "tinytex" "tippy" "TiPS"
[445] "tm" "tokenizers" "tools"
[448] "transformr" "translations" "tune"
[451] "tweenr" "tzdb" "UCSC.utils"
[454] "umap" "units" "urlchecker"
[457] "usa" "usdata" "usethis"
[460] "utf8" "utils" "uuid"
[463] "V8" "vcd" "vctrs"
[466] "venneuler" "viridis" "viridisLite"
[469] "visNetwork" "vroom" "waldo"
[472] "warp" "webr" "webshot"
[475] "webshot2" "websocket" "wesanderson"
[478] "whisker" "withr" "wk"
[481] "wordcloud" "workflows" "workflowsets"
[484] "writexl" "xfun" "XML"
[487] "xml2" "xopen" "xtable"
[490] "XVector" "yaml" "yardstick"
[493] "zip" "zoo" "ztable" Copying your setup
installed.packages() lists all installed packages
Search path
When you run library(pkg), the functions (and objects) in the package’s namespace are attached to the global search path.
[1] ".GlobalEnv" "package:stats" "package:graphics"
[4] "package:grDevices" "package:utils" "package:datasets"
[7] "package:methods" "Autoloads" "package:base" [1] ".GlobalEnv" "package:lubridate" "package:forcats"
[4] "package:stringr" "package:dplyr" "package:purrr"
[7] "package:readr" "package:tidyr" "package:tibble"
[10] "package:ggplot2" "package:tidyverse" "package:stats"
[13] "package:graphics" "package:grDevices" "package:utils"
[16] "package:datasets" "package:methods" "Autoloads"
[19] "package:base" Loading vs attaching
If you do not want to attach a package you can directly use package functions via ::
useful for avoiding namespace conflicts with functions of the same name from different packages.
requireNamespace()returnsTRUEorFALSEdepending on if it succeeds - can be useful to test if a package is available.
[1] "gtable" "jsonlite" "dplyr" "compiler" "stats"
[6] "tidyselect" "tidyverse" "stringr" "tidyr" "scales"
[11] "yaml" "fastmap" "base" "ggplot2" "readr"
[16] "R6" "generics" "knitr" "datasets" "methods"
[21] "forcats" "tibble" "lubridate" "pillar" "RColorBrewer"
[26] "tzdb" "rlang" "stringi" "xfun" "utils"
[31] "S7" "timechange" "cli" "withr" "magrittr"
[36] "digest" "grid" "rstudioapi" "graphics" "hms"
[41] "lifecycle" "vctrs" "evaluate" "glue" "farver"
[46] "rmarkdown" "purrr" "grDevices" "tools" "pkgconfig"
[51] "htmltools" Where do R packages come from?

Where do R packages come from?
- CRAN
- GitHub
- Local installation
From the terminal
or from R
Why TeachingDemos? See TeachingDemos::stork
What is CRAN?
The Comprehensive R Archive Network which is the central repository of R packages.
Maintained by the R Foundation and run by a team of volunteers, ~23k packages
Retains all current versions of released packages as well as archives of previous versions
Similar in spirit to Perl’s CPAN, TeX’s CTAN, and Python’s PyPI
Some important features:
All submissions are reviewed by humans + automated checks
Strictly enforced submission policies and package requirements
All packages must be actively maintained and support upstream and downstream changes
Structure of an R Package
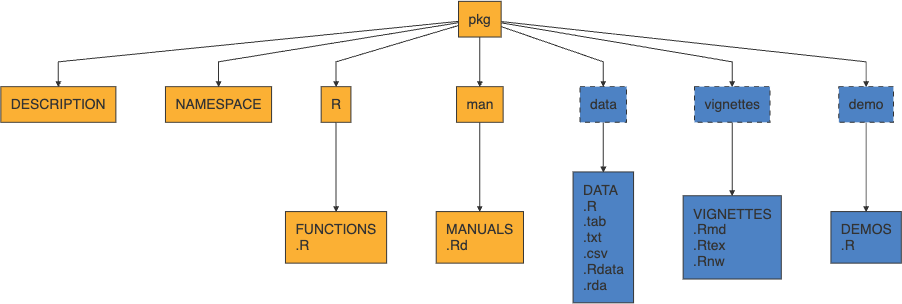
- yellow is core, blue is optional (but very common)
.Rdstands for “R documentation”
Core components
DESCRIPTION- file containing package metadata (e.g. package name, description, version, license, and author details). Also specifies package dependencies.NAMESPACE- details which functions and objects are exported by your package.R/- contains all R script files (.R) implementing packageman/- contains all R documentation files (.Rd)
Optional components
The following components are optional, but quite common:
tests/- contains unit tests (.Rscripts)src/- contains code to be compiled (usually C / C++)data/- contains example data sets (.rdsor.rda)inst/- contains files that will be copied to the package’s top-level directory when it is installed (e.g. C/C++ headers, examples or data files that don’t belong indata/)vignettes/- contains long form documentation, can be static (.pdfor.html) or literate documents (e.g..qmd,.Rmdor.Rnw)
Package contents
Installed Package
/Library/Frameworks/R.framework/Versions/4.5-arm64/Resources/library/TeachingDemos
├── DESCRIPTION
├── INDEX
├── Meta
│ ├── Rd.rds
│ ├── data.rds
│ ├── features.rds
│ ├── hsearch.rds
│ ├── links.rds
│ ├── nsInfo.rds
│ └── package.rds
├── NAMESPACE
├── NEWS
├── R
│ ├── TeachingDemos
│ ├── TeachingDemos.rdb
│ └── TeachingDemos.rdx
├── data
│ ├── Rdata.rdb
│ ├── Rdata.rds
│ └── Rdata.rdx
├── help
│ ├── AnIndex
│ ├── TeachingDemos.rdb
│ ├── TeachingDemos.rdx
│ ├── aliases.rds
│ └── paths.rds
└── html
├── 00Index.html
└── R.cssMake a package
Package development
What follows is an opinionated introduction to package development,
this is not the only way to make packages (none of the following are required)
I would strongly recommend using:
- RStudio
- RStudio projects
- GitHub
- usethis
- roxygen2
Read and follow along with R Packages (2e) - Chapter 1 - “The Whole Game”

usethis
This is an immensely useful package for automating all kinds of routine (and tedious) tasks within R
Tools for managing git and GitHub configuration
Tools for managing collaboration on GitHub via pull requests (see
pr_*())Tools for creating and configuring packages
Tools for configuring your R environment (e.g.
.Rprofileand.Renviron)and much much more
Note: the package also loads with devtools.
Live demo
Building a Package
Start your package
Rather than having to remember all of the necessary pieces and their format, usethis can help you bootstrap your package development process.
Choosing a license
An important early step in developing a package is choosing a license - this is not trivial but is important to do early on, particularly if collaborating with others.
There are many resources available to help you choose a license, including:
Creating a function for the package
Create a new .R file in the R folder with the function name as the title. E.g. use the Newton’s method code and call the file newton.
Documentation
All R packages are expected to have documentation for all exported functions and data sets (this is a CRAN requirement). This documentation is stored as .Rd files in the man/ directory.
The Rd format is a markup language that is loosely based on LaTeX
Rd files are processed into LaTeX, HTML, and plain text when building the package
All packages need Rd files, that doesn’t mean you need to write Rd
Roxygen2
The premise of roxygen2 is simple: describe your functions in comments next to their definitions and roxygen2 will process your source code and comments to automatically generate
.Rdfiles inman/,NAMESPACE, and, if needed, theCollatefield inDESCRIPTION.
roxygen uses special comment lines prefixed with
#'roxygen specific command have the format
@cmdand mostly matchRdcommandsdevtools::document()or menuBuild > Documentwith reprocess all source files and rebuild allRdsusethis::create_package()withroxygen = TRUEwill initialize your package to use roxygen (default behavior)
Example
#' newton
#'
#' This function takes four arguments: f, fp, x, tol and returns the root a root of f. Note that this function is sensitive to the starting point x.
#'
#'
#' @param f univariate function that you want to find the root of.
#' @param fp first derivative of function f in closed form.
#' @param x starting point
#' @param tol The tolerance term, i.e. how close the function must be to zero in order to terminate.
#' @return Returns an object of indeterminate type because the function is not robustly written.
#' @exportRun devtools::document().
Check out ?newton
Package vigenette(s)
Vignette
Long form documentation for your package that live in vignette/, use browseVignette(pkg) to see a package’s vignettes.
Not required, but adds a lot of value to a package
Generally these are literate documents (
.Rmd,.Rnw) that are compiled to.htmlor.pdfwhen the package is built.Built packages retain the rendered document, the source document, and all source code
vignette("colwise", package = "dplyr")opens rendered versionedit(vignette("colwise", package = "dplyr"))opens code chunks
Use
usethis::use_vignette()to create a RMarkdown vignette template
Articles
These are un-official extensions to vignettes where package authors wish to include additional long form documentation that is included in their pkgdown site but not in the package (usually for space reasons).
Use
usethis::use_article()to createFiles are added to
vignette/articles/which is added to.Rbuildignore.Rbuildignoreexcludes certain files from the package build process # Package data
Exported data
Many packages contain sample data (e.g. nycflights13, babynames, etc.)
Generally these files are made available by saving a single data object as an .Rdata file (using save()) into the data/ directory of your package.
An easy option is to use
usethis::use_data(obj)to create the necessary file(s)Data is usually compressed, for large data sets it may be worth trying different options (there is a 5 Mb package size limit on CRAN)
Exported data must be documented (possible via roxygen)
Lazy data
By default when attaching a package all of that packages data is loaded - however if LazyData: true is set in the packages’ DESCRIPTION then data is only loaded when used.
Raw data
When published, a package should generally only contain the final data set, but it is important that the process to generate the data is documented as well as any necessary preliminary data.
These can live any where but the general suggestion is to create a
data-raw/directory which is ignored via.Rbuildignoredata-raw/then contain scripts, data files, and anything else needed to generate the final objectSee examples babynames or nycflights
Use
usethis::use_data_raw()to create and ignore thedata-raw/directory.
Internal data
If you have data that you want to have access to from within the package but not exported then it needs to live in a special Rdata object located at R/sysdata.rda.
Can be created using
usethis::use_data(obj1, obj2, internal = TRUE)Each call to the above will overwrite, so needs to include all objects
Not necessary for small data frames and similar objects - just create in a script. Use when you want the object to be compressed.
Example nflplotR which contains team logos and colors for NFL teams.
Raw data files
If you want to include raw data files (e.g .csv, shapefiles, etc.) there are generally placed in inst/ (or a nested folder) so that they are installed with the package.
Accessed using
system.file("dir", package = "package")after installUse folders to keep things organized, Hadley recommends and uses
inst/extdata/Example sf
Package checking
R CMD check
Last time we saw the usage of R CMD check, or rather Build > Check Package from within RStudio.
This is a good idea to run regularly to make sure nothing is broken and you are meeting the important package quality standards, but this only in the context of your machine, your version of R, your OS, and so on.
If using GitHub it is highly recommended that you run usethis::use_github_action_check_standard() to enable GitHub actions checks of the package each time it is pushed.
On each push this runs R CMD check on: * Latest R on MacOS, Windows, Linux (Ubuntu) * Previous and development version of R on Linux (Ubuntu)
Package testing
Basic test structure
Package tests live in tests/,
Any R scripts found in the folder will be run when checking the package (not Building)
Generally tests fail on errors, but warnings are also tracked
Testing is possible via base R, including comparison of output vs. a file but it is not recommended (See Writing R Extensions)
Note that R CMD check also runs all documentation examples (unless explicitly tagged dont run) - which can be used for basic testing
testthat basics
Not the only option but probably the most widely used and with the best integration into RStudio.
Can be initialized in your project via usethis::use_testthat() which creates tests/testthat/ and some basic scaffolding.
test/testthat.Ris what is run by R CMD Check and runs your other tests - handles some basic config like loading package(s)Test scripts go in
tests/testthat/and should start withtest_, suffix is usually the file inR/that is being tested.
testthat script structure
From the bottom up,
a single test is written as an expectation (e.q.
expect_equal(),expect_error(), etc.)multiple related expectations are combined into a test group (
test_that()), which provides- a human readable name and
- local scope to contain the expectations and any temporary objects
multiple test groups are combined into a file
Installing the package
How to
After building, you will see package_name_version.tar.gz appear in the parent directory.
We can install with